ribs.archives.CVTArchive¶
- class ribs.archives.CVTArchive(*, solution_dim, cells, ranges, learning_rate=None, threshold_min=-inf, qd_score_offset=0.0, seed=None, dtype=<class 'numpy.float64'>, extra_fields=None, custom_centroids=None, centroid_method='kmeans', samples=100000, k_means_kwargs=None, use_kd_tree=True, ckdtree_kwargs=None, chunk_size=None)[source]¶
An archive that divides the entire measure space into a fixed number of cells.
This archive originates in Vassiliades 2018. It uses Centroidal Voronoi Tessellation (CVT) to divide an n-dimensional measure space into k cells. The CVT is created by sampling points uniformly from the n-dimensional measure space and using k-means clustering to identify k centroids. When items are inserted into the archive, we identify their cell by identifying the closest centroid in measure space (using Euclidean distance). For k-means clustering, we use
sklearn.cluster.k_means().By default, finding the closest centroid is done in roughly O(log(number of cells)) time using
scipy.spatial.cKDTree. To switch to brute force, which takes O(number of cells) time, passuse_kd_tree=False.To compare the performance of using the k-D tree vs brute force, we ran benchmarks where we inserted 1k batches of 100 solutions into a 2D archive with varying numbers of cells. We took the minimum over 5 runs for each data point, as recommended in the docs for
timeit.Timer.repeat(). Note the logarithmic scales. This plot was generated on a reasonably modern laptop.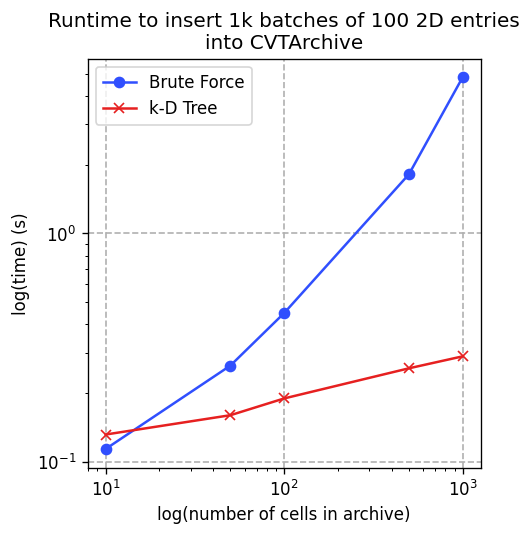
Across almost all numbers of cells, using the k-D tree is faster than using brute force. Thus, we recommend always using the k-D tree. See benchmarks/cvt_add.py in the project repo for more information about how this plot was generated.
Finally, if running multiple experiments, it may be beneficial to use the same centroids across each experiment. Doing so can keep experiments consistent and reduce execution time. To do this, either (1) construct custom centroids and pass them in via the
custom_centroidsargument, or (2) access the centroids created in the first archive withcentroidsand pass them intocustom_centroidswhen constructing archives for subsequent experiments.Note
The idea of archive thresholds was introduced in Fontaine 2022. For more info on thresholds, including the
learning_rateandthreshold_minparameters, refer to our tutorial Upgrading CMA-ME to CMA-MAE on the Sphere Benchmark.Note
For more information on our choice of k-D tree implementation, see #38.
- Parameters
solution_dim (int) – Dimension of the solution space.
cells (int) – The number of cells to use in the archive, equivalent to the number of centroids/areas in the CVT.
ranges (array-like of (float, float)) – Upper and lower bound of each dimension of the measure space, e.g.
[(-1, 1), (-2, 2)]indicates the first dimension should have bounds \([-1,1]\) (inclusive), and the second dimension should have bounds \([-2,2]\) (inclusive).rangesshould be the same length asdims.learning_rate (float) – The learning rate for threshold updates. Defaults to 1.0.
threshold_min (float) – The initial threshold value for all the cells.
qd_score_offset (float) – Archives often contain negative objective values, and if the QD score were to be computed with these negative objectives, the algorithm would be penalized for adding new cells with negative objectives. Thus, a standard practice is to normalize all the objectives so that they are non-negative by introducing an offset. This QD score offset will be subtracted from all objectives in the archive, e.g., if your objectives go as low as -300, pass in -300 so that each objective will be transformed as
objective - (-300).seed (int) – Value to seed the random number generator as well as
k_means(). Set to None to avoid a fixed seed.dtype (str or data-type) – Data type of the solutions, objectives, and measures. We only support
"f"/np.float32and"d"/np.float64.extra_fields (dict) – Description of extra fields of data that is stored next to elite data like solutions and objectives. The description is a dict mapping from a field name (str) to a tuple of
(shape, dtype). For instance,{"foo": ((), np.float32), "bar": ((10,), np.float32)}will create a “foo” field that contains scalar values and a “bar” field that contains 10D values. Note that field names must be valid Python identifiers, and names already used in the archive are not allowed.custom_centroids (array-like) – If passed in, this (cells, measure_dim) array will be used as the centroids of the CVT instead of generating new ones. In this case,
sampleswill be ignored, andarchive.sampleswill be None. This can be useful when one wishes to use the same CVT across experiments for fair comparison.centroid_method (str) – Pass in the following methods for generating centroids: “random”, “sobol”, “scrambled_sobol”, “halton”. Default method is “kmeans”. These methods are derived from Mouret 2023: https://dl.acm.org/doi/pdf/10.1145/3583133.3590726. Note: Samples are only used when method is “kmeans”.
samples (int or array-like) – If it is an int, this specifies the number of samples to generate when creating the CVT. Otherwise, this must be a (num_samples, measure_dim) array where samples[i] is a sample to use when creating the CVT. It can be useful to pass in custom samples when there are restrictions on what samples in the measure space are (physically) possible.
k_means_kwargs (dict) – kwargs for
k_means(). By default, we pass in n_init=1, init=”random”, algorithm=”lloyd”, and random_state=seed.use_kd_tree (bool) – If True, use a k-D tree for finding the closest centroid when inserting into the archive. If False, brute force will be used instead.
ckdtree_kwargs (dict) – kwargs for
cKDTree. By default, we do not pass in any kwargs.chunk_size (int) – If passed, brute forcing the closest centroid search will chunk the distance calculations to compute chunk_size inputs at a time.
- Raises
ValueError – The
samplesarray or thecustom_centroidsarray has the wrong shape.
Methods
__iter__()Creates an iterator over the elites in the archive.
__len__()Number of elites in the archive.
add(solution, objective, measures, **fields)Inserts a batch of solutions into the archive.
add_single(solution, objective, measures, ...)Inserts a single solution into the archive.
as_pandas([include_solutions, include_metadata])DEPRECATED.
clear()Removes all elites from the archive.
cqd_score(iterations, target_points, ...[, ...])Computes the CQD score of the archive.
data([fields, return_type])Retrieves data for all elites in the archive.
index_of(measures)Finds the indices of the centroid closest to the given coordinates in measure space.
index_of_single(measures)Returns the index of the measures for one solution.
retrieve(measures)Retrieves the elites with measures in the same cells as the measures specified.
retrieve_single(measures)Retrieves the elite with measures in the same cell as the measures specified.
Randomly samples elites from the archive.
Attributes
The elite with the highest objective in the archive.
Total number of cells in the archive.
The centroids used in the CVT.
The dtype of the solutions, objective, and measures.
Whether the archive is empty.
List of data fields in the archive.
The size of each dim (upper_bounds - lower_bounds).
The learning rate for threshold updates.
Lower bound of each dimension.
Dimensionality of the measure space.
The offset which is subtracted from objective values when computing the QD score.
The samples used in creating the CVT.
Dimensionality of the solutions in the archive.
Statistics about the archive.
The initial threshold value for all the cells.
Upper bound of each dimension.
- __iter__()¶
Creates an iterator over the elites in the archive.
Example
for elite in archive: elite["solution"] elite["objective"] ...
- __len__()¶
Number of elites in the archive.
- add(solution, objective, measures, **fields)¶
Inserts a batch of solutions into the archive.
Each solution is only inserted if it has a higher
objectivethan the threshold of the corresponding cell. For the default values oflearning_rateandthreshold_min, this threshold is simply the objective value of the elite previously in the cell. If multiple solutions in the batch end up in the same cell, we only insert the solution with the highest objective. If multiple solutions end up in the same cell and tie for the highest objective, we insert the solution that appears first in the batch.For the default values of
learning_rateandthreshold_min, the threshold for each cell is updated by taking the maximum objective value among all the solutions that landed in the cell, resulting in the same behavior as in the vanilla MAP-Elites archive. However, for other settings, the threshold is updated with the batch update rule described in the appendix of Fontaine 2022.Note
The indices of all arguments should “correspond” to each other, i.e.
solution[i],objective[i],measures[i], and should be the solution parameters, objective, and measures for solutioni.- Parameters
solution (array-like) – (batch_size,
solution_dim) array of solution parameters.objective (array-like) – (batch_size,) array with objective function evaluations of the solutions.
measures (array-like) – (batch_size,
measure_dim) array with measure space coordinates of all the solutions.fields (keyword arguments) – Additional data for each solution. Each argument should be an array with batch_size as the first dimension.
- Returns
Information describing the result of the add operation. The dict contains the following keys:
"status"(numpy.ndarrayofint): An array of integers that represent the “status” obtained when attempting to insert each solution in the batch. Each item has the following possible values:0: The solution was not added to the archive.1: The solution improved the objective value of a cell which was already in the archive.2: The solution discovered a new cell in the archive.
All statuses (and values, below) are computed with respect to the current archive. For example, if two solutions both introduce the same new archive cell, then both will be marked with
2.The alternative is to depend on the order of the solutions in the batch – for example, if we have two solutions
aandbwhich introduce the same new cell in the archive,acould be inserted first with status2, andbcould be inserted second with status1because it improves upona. However, our implementation does not do this.To convert statuses to a more semantic format, cast all statuses to
AddStatuse.g. with[AddStatus(s) for s in add_info["status"]]."value"(numpy.ndarrayofdtype): An array with values for each solution in the batch. With the default values oflearning_rate = 1.0andthreshold_min = -np.inf, the meaning of each value depends on the correspondingstatusand is identical to that in CMA-ME (Fontaine 2020):0(not added): The value is the “negative improvement,” i.e. the objective of the solution passed in minus the objective of the elite still in the archive (this value is negative because the solution did not have a high enough objective to be added to the archive).1(improve existing cell): The value is the “improvement,” i.e. the objective of the solution passed in minus the objective of the elite previously in the archive.2(new cell): The value is just the objective of the solution.
In contrast, for other values of
learning_rateandthreshold_min, each value is equivalent to the objective value of the solution minus the threshold of its corresponding cell in the archive.
- Return type
- Raises
ValueError – The array arguments do not match their specified shapes.
ValueError –
objectiveormeasureshas non-finite values (inf or NaN).
- add_single(solution, objective, measures, **fields)¶
Inserts a single solution into the archive.
The solution is only inserted if it has a higher
objectivethan the threshold of the corresponding cell. For the default values oflearning_rateandthreshold_min, this threshold is simply the objective value of the elite previously in the cell. The threshold is also updated if the solution was inserted.Note
To make it more amenable to modifications, this method’s implementation is designed to be readable at the cost of performance, e.g., none of its operations are modified. If you need performance, we recommend using
add().- Parameters
solution (array-like) – Parameters of the solution.
objective (float) – Objective function evaluation of the solution.
measures (array-like) – Coordinates in measure space of the solution.
fields (keyword arguments) – Additional data for the solution.
- Returns
Information describing the result of the add operation. The dict contains
statusandvaluekeys; refer toadd()for the meaning of status and value.- Return type
- Raises
ValueError – The array arguments do not match their specified shapes.
ValueError –
objectiveis non-finite (inf or NaN) ormeasureshas non-finite values.
- as_pandas(include_solutions=True, include_metadata=False)¶
DEPRECATED.
- clear()¶
Removes all elites from the archive.
After this method is called, the archive will be
empty.
- cqd_score(iterations, target_points, penalties, obj_min, obj_max, dist_max=None, dist_ord=None)¶
Computes the CQD score of the archive.
The Continuous Quality Diversity (CQD) score was introduced in Kent 2022.
Note
This method by default assumes that the archive has an
upper_boundsandlower_boundsproperty which delineate the bounds of the measure space, as is the case inGridArchive,CVTArchive, andSlidingBoundariesArchive. If this is not the case,dist_maxmust be passed in, andtarget_pointsmust be an array of custom points.- Parameters
iterations (int) – Number of times to compute the CQD score. We return the mean CQD score across these iterations.
target_points (int or array-like) – Number of target points to generate, or an (iterations, n, measure_dim) array which lists n target points to list on each iteration. When an int is passed, the points are sampled uniformly within the bounds of the measure space.
penalties (int or array-like) – Number of penalty values over which to compute the score (the values are distributed evenly over the range [0,1]). Alternatively, this may be a 1D array which explicitly lists the penalty values. Known as \(\theta\) in Kent 2022.
obj_min (float) – Minimum objective value, used when normalizing the objectives.
obj_max (float) – Maximum objective value, used when normalizing the objectives.
dist_max (float) – Maximum distance between points in measure space. Defaults to the distance between the extremes of the measure space bounds (the type of distance is computed with the order specified by
dist_ord). Known as \(\delta_{max}\) in Kent 2022.dist_ord – Order of the norm to use for calculating measure space distance; this is passed to
numpy.linalg.norm()as theordargument. Seenumpy.linalg.norm()for possible values. The default is to use Euclidean distance (L2 norm).
- Returns
The mean CQD score obtained with
iterationsrounds of calculations.- Raises
RuntimeError – The archive does not have the bounds properties mentioned above, and dist_max is not specified or the target points are not provided.
ValueError – target_points or penalties is an array with the wrong shape.
- data(fields=None, return_type='dict')¶
Retrieves data for all elites in the archive.
- Parameters
fields (str or array-like of str) – List of fields to include. By default, all fields will be included, with an additional “index” as the last field (“index” can also be placed anywhere in this list). This can also be a single str indicating a field name.
return_type (str) – Type of data to return. See below. Ignored if
fieldsis a str.
- Returns
The data for all entries in the archive. If
fieldswas a single str, this will just be an array holding data for the given field. Otherwise, this data can take the following forms, depending on thereturn_typeargument:return_type="dict": Dict mapping from the field name to the field data at the given indices. An example is:{ "solution": [[1.0, 1.0, ...], ...], "objective": [1.5, ...], "measures": [[1.0, 2.0], ...], "threshold": [0.8, ...], "index": [4, ...], }
Observe that we also return the indices as an
indexentry in the dict. The keys in this dict can be modified with thefieldsarg; duplicate fields will be ignored since the dict stores unique keys.return_type="tuple": Tuple of arrays matching the field order given infields. For instance, iffieldswas["objective", "measures"], we would receive a tuple of(objective_arr, measures_arr). In this case, the results fromretrievecould be unpacked as:objective, measures = archive.data(["objective", "measures"], return_type="tuple")
Unlike with the
dictreturn type, duplicate fields will show up as duplicate entries in the tuple, e.g.,fields=["objective", "objective"]will result in two objective arrays being returned.By default, (i.e., when
fields=None), the fields in the tuple will be ordered according to thefield_listalong withindexas the last field.return_type="pandas": AArchiveDataFramewith the following columns:For fields that are scalars, a single column with the field name. For example,
objectivewould have a single column calledobjective.For fields that are 1D arrays, multiple columns with the name suffixed by its index. For instance, if we have a
measuresfield of length 10, we create 10 columns with namesmeasures_0,measures_1, …,measures_9. We do not currently support fields with >1D data.1 column of integers (
np.int32) for the index, namedindex.
In short, the dataframe might look like this by default:
solution_0
…
objective
measures_0
…
threshold
index
…
…
Like the other return types, the columns can be adjusted with the
fieldsparameter.
All data returned by this method will be a copy, i.e., the data will not update as the archive changes.
- index_of(measures)[source]¶
Finds the indices of the centroid closest to the given coordinates in measure space.
If
index_batchis the batch of indices returned by this method, thenarchive.centroids[index_batch[i]]holds the coordinates of the centroid closest tomeasures[i]. Seecentroidsfor more info.The centroid indices are located using either the k-D tree or brute force, depending on the value of
use_kd_treein the constructor.- Parameters
measures (array-like) – (batch_size,
measure_dim) array of coordinates in measure space.- Returns
(batch_size,) array of centroid indices corresponding to each measure space coordinate.
- Return type
- Raises
ValueError –
measuresis not of shape (batch_size,measure_dim).ValueError –
measureshas non-finite values (inf or NaN).
- index_of_single(measures)¶
Returns the index of the measures for one solution.
While
index_of()takes in a batch of measures, this method takes in the measures for only one solution. Ifindex_of()is implemented correctly, this method should work immediately (i.e. “out of the box”).- Parameters
measures (array-like) – (
measure_dim,) array of measures for a single solution.- Returns
Integer index of the measures in the archive’s storage arrays.
- Return type
int or numpy.integer
- Raises
ValueError –
measuresis not of shape (measure_dim,).ValueError –
measureshas non-finite values (inf or NaN).
- retrieve(measures)¶
Retrieves the elites with measures in the same cells as the measures specified.
This method operates in batch, i.e., it takes in a batch of measures and outputs the batched data for the elites:
occupied, elites = archive.retrieve(...) elites["solution"] # Shape: (batch_size, solution_dim) elites["objective"] elites["measures"] elites["threshold"] elites["index"]
If the cell associated with
elites["measures"][i]has an elite in it, thenoccupied[i]will be True. Furthermore,elites["solution"][i],elites["objective"][i],elites["measures"][i],elites["threshold"][i], andelites["index"][i]will be set to the properties of the elite. Note thatelites["measures"][i]may not be equal to themeasures[i]passed as an argument, since the measures only need to be in the same archive cell.If the cell associated with
measures[i]does not have any elite in it, thenoccupied[i]will be set to False. Furthermore, the corresponding outputs will be set to empty values – namely:NaN for floating-point fields
-1 for the “index” field
0 for integer fields
None for object fields
If you need to retrieve a single elite associated with some measures, consider using
retrieve_single().- Parameters
measures (array-like) – (batch_size,
measure_dim) array of coordinates in measure space.- Returns
2-element tuple of (occupied array, dict). The occupied array indicates whether each of the cells indicated by the coordinates in
measureshas an elite, while the dict contains the data of those elites. The dict maps from field name to the corresponding array.- Return type
- Raises
ValueError –
measuresis not of shape (batch_size,measure_dim).ValueError –
measureshas non-finite values (inf or NaN).
- retrieve_single(measures)¶
Retrieves the elite with measures in the same cell as the measures specified.
While
retrieve()takes in a batch of measures, this method takes in the measures for only one solution and returns a single bool and a dict with single entries.- Parameters
measures (array-like) – (
measure_dim,) array of measures.- Returns
If there is an elite with measures in the same cell as the measures specified, then this method returns a True value and a dict where all the fields hold the info of the elite. Otherwise, this method returns a False value and a dict filled with the same “empty” values described in
retrieve().- Return type
- Raises
ValueError –
measuresis not of shape (measure_dim,).ValueError –
measureshas non-finite values (inf or NaN).
- sample_elites(n)¶
Randomly samples elites from the archive.
Currently, this sampling is done uniformly at random. Furthermore, each sample is done independently, so elites may be repeated in the sample. Additional sampling methods may be supported in the future.
Example
elites = archive.sample_elites(16) elites["solution"] # Shape: (16, solution_dim) elites["objective"] ...
- Parameters
n (int) – Number of elites to sample.
- Returns
Holds a batch of elites randomly selected from the archive.
- Return type
- Raises
IndexError – The archive is empty.
- property best_elite¶
The elite with the highest objective in the archive.
None if there are no elites in the archive.
Note
If the archive is non-elitist (this occurs when using the archive with a learning rate which is not 1.0, as in CMA-MAE), then this best elite may no longer exist in the archive because it was replaced with an elite with a lower objective value. This can happen because in non-elitist archives, new solutions only need to exceed the threshold of the cell they are being inserted into, not the objective of the elite currently in the cell. See #314 for more info.
Note
The best elite will contain a “threshold” key. This threshold is the threshold of the best elite’s cell after the best elite was inserted into the archive.
- Type
- property centroids¶
The centroids used in the CVT.
- Type
(n_centroids, measure_dim) numpy.ndarray
- property dtype¶
The dtype of the solutions, objective, and measures.
- Type
data-type
- property interval_size¶
The size of each dim (upper_bounds - lower_bounds).
- Type
(measure_dim,) numpy.ndarray
- property lower_bounds¶
Lower bound of each dimension.
- Type
(measure_dim,) numpy.ndarray
- property qd_score_offset¶
The offset which is subtracted from objective values when computing the QD score.
- Type
- property samples¶
The samples used in creating the CVT.
Will be None if custom centroids were passed in to the archive.
- Type
(num_samples, measure_dim) numpy.ndarray
- property stats¶
Statistics about the archive.
See
ArchiveStatsfor more info.- Type
- property upper_bounds¶
Upper bound of each dimension.
- Type
(measure_dim,) numpy.ndarray